Identification of Predominant Histopathological Growth Patterns of Colorectal Liver Metastasis by Multi-Habitat and Multi-Sequence Based Radiomics Analysis
- PMID: 32923388
- PMCID: PMC7456817
- DOI: 10.3389/fonc.2020.01363
Identification of Predominant Histopathological Growth Patterns of Colorectal Liver Metastasis by Multi-Habitat and Multi-Sequence Based Radiomics Analysis
Abstract
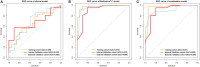
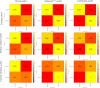
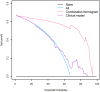
Purpose: Developing an MRI-based radiomics model to effectively and accurately predict the predominant histopathologic growth patterns (HGPs) of colorectal liver metastases (CRLMs). Materials and Methods: In this study, 182 resected and histopathological proven CRLMs of chemotherapy-naive patients from two institutions, including 123 replacement CRLMs and 59 desmoplastic CRLMs, were retrospectively analyzed. Radiomics analysis was performed on two regions of interest (ROI), the tumor zone and the tumor-liver interface (TLI) zone. Decision tree (DT) algorithm was used for radiomics modeling on each MR sequence, and fused radiomics model was constructed by combining the radiomics signature of each sequence. The clinical and combination models were developed through multivariate logistic regression method. The performance of the developed models was assessed by receiver operating characteristic (ROC) curves with indicators of area under curve (AUC), accuracy, sensitivity, and specificity. A nomogram was constructed to evaluate the discrimination, calibration, and usefulness. Results: The fused radiomicstumor and radiomicsTLI models showed better performance than any single sequence and clinical model. In addition, the radiomicsTLI model exhibited better performance than radiomicstumor model (AUC of 0.912 vs. 0.879) in internal validation cohort. The combination model showed good discrimination, and the AUC of nomogram was 0.971, 0.909, and 0.905 in the training, internal validation, and external validation cohorts, respectively. Conclusion: MRI-based radiomics method has high potential in predicting the predominant HGPs of CRLM. Preoperative non-invasive identification of predominant HGPs could further explore the ability of HGPs as a potential biomarker for clinical treatment strategy, reflecting different biological pathways.
Keywords: colorectal cancer; histopathologic growth patterns; liver metastasis; magnetic resonance; radiomics.
Copyright © 2020 Han, Chai, Wei, Yue, Cheng, Gu, Zhang, Tong, Sheng, Hong, Ye, Wang and Tian.
Figures



Similar articles
-
CT, MRI, and radiomics studies of liver metastasis histopathological growth patterns: an up-to-date review.Abdom Radiol (NY). 2022 Oct;47(10):3494-3506. doi: 10.1007/s00261-022-03616-z. Epub 2022 Jul 27. Abdom Radiol (NY). 2022. PMID: 35895118 Review.
-
Prediction of Histopathologic Growth Patterns of Colorectal Liver Metastases with a Noninvasive Imaging Method.Ann Surg Oncol. 2019 Dec;26(13):4587-4598. doi: 10.1245/s10434-019-07910-x. Epub 2019 Oct 11. Ann Surg Oncol. 2019. PMID: 31605342
-
Pre-operative prediction of histopathological growth patterns of colorectal cancer liver metastasis using MRI-based radiomic models.Abdom Radiol (NY). 2024 Dec;49(12):4239-4248. doi: 10.1007/s00261-024-04290-z. Epub 2024 Jul 29. Abdom Radiol (NY). 2024. PMID: 39069557
-
Prediction of transformation in the histopathological growth pattern of colorectal liver metastases after chemotherapy using CT-based radiomics.Clin Exp Metastasis. 2024 Apr;41(2):143-154. doi: 10.1007/s10585-024-10275-5. Epub 2024 Feb 28. Clin Exp Metastasis. 2024. PMID: 38416301
-
Histopathological growth patterns of liver metastasis: updated consensus guidelines for pattern scoring, perspectives and recent mechanistic insights.Br J Cancer. 2022 Oct;127(6):988-1013. doi: 10.1038/s41416-022-01859-7. Epub 2022 Jun 1. Br J Cancer. 2022. PMID: 35650276 Free PMC article. Review.
Cited by
-
Clinical implications and mechanism of histopathological growth pattern in colorectal cancer liver metastases.World J Gastroenterol. 2022 Jul 14;28(26):3101-3115. doi: 10.3748/wjg.v28.i26.3101. World J Gastroenterol. 2022. PMID: 36051338 Free PMC article. Review.
-
A CT-based radiomics nomogram for predicting histopathologic growth patterns of colorectal liver metastases.J Cancer Res Clin Oncol. 2023 Sep;149(12):9543-9555. doi: 10.1007/s00432-023-04852-6. Epub 2023 May 23. J Cancer Res Clin Oncol. 2023. PMID: 37221440
-
The Histopathological Growth Pattern of Colorectal Liver Metastases Impacts Local Recurrence Risk and the Adequate Width of the Surgical Margin.Ann Surg Oncol. 2022 Sep;29(9):5515-5524. doi: 10.1245/s10434-022-11717-8. Epub 2022 Jun 10. Ann Surg Oncol. 2022. PMID: 35687176
-
CT, MRI, and radiomics studies of liver metastasis histopathological growth patterns: an up-to-date review.Abdom Radiol (NY). 2022 Oct;47(10):3494-3506. doi: 10.1007/s00261-022-03616-z. Epub 2022 Jul 27. Abdom Radiol (NY). 2022. PMID: 35895118 Review.
-
The histological growth patterns in liver metastases from colorectal cancer display differences in lymphoid, myeloid, and mesenchymal cells.MedComm (2020). 2024 Nov 19;5(12):e70000. doi: 10.1002/mco2.70000. eCollection 2024 Dec. MedComm (2020). 2024. PMID: 39563958 Free PMC article.
References
LinkOut - more resources
Full Text Sources

