Estimating the overdispersion in COVID-19 transmission using outbreak sizes outside China
- PMID: 32685698
- PMCID: PMC7338915
- DOI: 10.12688/wellcomeopenres.15842.3
Estimating the overdispersion in COVID-19 transmission using outbreak sizes outside China
Abstract
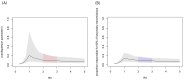
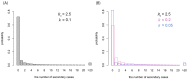
Background: A novel coronavirus disease (COVID-19) outbreak has now spread to a number of countries worldwide. While sustained transmission chains of human-to-human transmission suggest high basic reproduction number R 0, variation in the number of secondary transmissions (often characterised by so-called superspreading events) may be large as some countries have observed fewer local transmissions than others. Methods: We quantified individual-level variation in COVID-19 transmission by applying a mathematical model to observed outbreak sizes in affected countries. We extracted the number of imported and local cases in the affected countries from the World Health Organization situation report and applied a branching process model where the number of secondary transmissions was assumed to follow a negative-binomial distribution. Results: Our model suggested a high degree of individual-level variation in the transmission of COVID-19. Within the current consensus range of R 0 (2-3), the overdispersion parameter k of a negative-binomial distribution was estimated to be around 0.1 (median estimate 0.1; 95% CrI: 0.05-0.2 for R0 = 2.5), suggesting that 80% of secondary transmissions may have been caused by a small fraction of infectious individuals (~10%). A joint estimation yielded likely ranges for R 0 and k (95% CrIs: R 0 1.4-12; k 0.04-0.2); however, the upper bound of R 0 was not well informed by the model and data, which did not notably differ from that of the prior distribution. Conclusions: Our finding of a highly-overdispersed offspring distribution highlights a potential benefit to focusing intervention efforts on superspreading. As most infected individuals do not contribute to the expansion of an epidemic, the effective reproduction number could be drastically reduced by preventing relatively rare superspreading events.
Keywords: COVID-19; SARS-CoV-2; branching process; novel coronavirus; overdispersion; superspreading.
Copyright: © 2020 Endo A et al.
Conflict of interest statement
Competing interests: AE received a research grant from Taisho Pharmaceutical Co., Ltd.
Figures
Similar articles
-
Depressing time: Waiting, melancholia, and the psychoanalytic practice of care.In: Kirtsoglou E, Simpson B, editors. The Time of Anthropology: Studies of Contemporary Chronopolitics. Abingdon: Routledge; 2020. Chapter 5. In: Kirtsoglou E, Simpson B, editors. The Time of Anthropology: Studies of Contemporary Chronopolitics. Abingdon: Routledge; 2020. Chapter 5. PMID: 36137063 Free Books & Documents. Review.
-
Falls prevention interventions for community-dwelling older adults: systematic review and meta-analysis of benefits, harms, and patient values and preferences.Syst Rev. 2024 Nov 26;13(1):289. doi: 10.1186/s13643-024-02681-3. Syst Rev. 2024. PMID: 39593159 Free PMC article.
-
Comparison of Two Modern Survival Prediction Tools, SORG-MLA and METSSS, in Patients With Symptomatic Long-bone Metastases Who Underwent Local Treatment With Surgery Followed by Radiotherapy and With Radiotherapy Alone.Clin Orthop Relat Res. 2024 Dec 1;482(12):2193-2208. doi: 10.1097/CORR.0000000000003185. Epub 2024 Jul 23. Clin Orthop Relat Res. 2024. PMID: 39051924
-
Ceftazidime with avibactam for treating severe aerobic Gram-negative bacterial infections: technology evaluation to inform a novel subscription-style payment model.Health Technol Assess. 2024 Oct;28(73):1-230. doi: 10.3310/YAPL9347. Health Technol Assess. 2024. PMID: 39487661 Free PMC article.
-
Trends in Surgical and Nonsurgical Aesthetic Procedures: A 14-Year Analysis of the International Society of Aesthetic Plastic Surgery-ISAPS.Aesthetic Plast Surg. 2024 Oct;48(20):4217-4227. doi: 10.1007/s00266-024-04260-2. Epub 2024 Aug 5. Aesthetic Plast Surg. 2024. PMID: 39103642 Review.
Cited by
-
Slight reduction in SARS-CoV-2 exposure viral load due to masking results in a significant reduction in transmission with widespread implementation.Sci Rep. 2021 Jun 4;11(1):11838. doi: 10.1038/s41598-021-91338-5. Sci Rep. 2021. PMID: 34088959 Free PMC article.
-
Just 2% of SARS-CoV-2-positive individuals carry 90% of the virus circulating in communities.Proc Natl Acad Sci U S A. 2021 May 25;118(21):e2104547118. doi: 10.1073/pnas.2104547118. Proc Natl Acad Sci U S A. 2021. PMID: 33972412 Free PMC article.
-
Stay-At-Home Orders Are Associated With Emergence of Novel SARS-CoV-2 Variants.Cureus. 2021 Mar 11;13(3):e13819. doi: 10.7759/cureus.13819. Cureus. 2021. PMID: 33728228 Free PMC article.
-
The role of inter-regional mobility in forecasting SARS-CoV-2 transmission.J R Soc Interface. 2022 Aug;19(193):20220486. doi: 10.1098/rsif.2022.0486. Epub 2022 Aug 31. J R Soc Interface. 2022. PMID: 36043288 Free PMC article.
-
Using a household-structured branching process to analyse contact tracing in the SARS-CoV-2 pandemic.Philos Trans R Soc Lond B Biol Sci. 2021 Jul 19;376(1829):20200267. doi: 10.1098/rstb.2020.0267. Epub 2021 May 31. Philos Trans R Soc Lond B Biol Sci. 2021. PMID: 34053253 Free PMC article.
References
-
- Abbott S, Hellewell J, Munday J, et al. : The transmissibility of novel Coronavirus in the early stages of the 2019-20 outbreak in Wuhan: Exploring initial point-source exposure sizes and durations using scenario analysis [version 1; peer review: 1 approved]. Wellcome Open Res. 2020;5:17. 10.12688/wellcomeopenres.15718.1 - DOI - PMC - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

