A rotary plasmonic nanoclock
- PMID: 31776340
- PMCID: PMC6881389
- DOI: 10.1038/s41467-019-13444-3
A rotary plasmonic nanoclock
Abstract
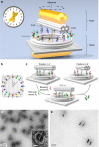
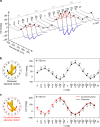
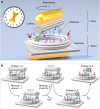
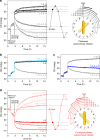
One of the fundamental challenges in nanophotonics is to gain full control over nanoscale optical elements. The precise spatiotemporal arrangement determines their interactions and collective behavior. To this end, DNA nanotechnology is employed as an unprecedented tool to create nanophotonic devices with excellent spatial addressability and temporal programmability. However, most of the current DNA-assembled nanophotonic devices can only reconfigure among random or very few defined states. Here, we demonstrate a DNA-assembled rotary plasmonic nanoclock. In this system, a rotor gold nanorod can carry out directional and reversible 360° rotation with respect to a stator gold nanorod, transitioning among 16 well-defined configurations powered by DNA fuels. The full-turn rotation process is monitored by optical spectroscopy in real time. We further demonstrate autonomous rotation of the plasmonic nanoclock powered by DNAzyme-RNA interactions. Such assembly approaches pave a viable route towards advanced nanophotonic systems entirely from the bottom-up.
Conflict of interest statement
The authors declare no competing interests.
Figures
Similar articles
-
DNA-Assembled Multilayer Sliding Nanosystems.Nano Lett. 2019 Sep 11;19(9):6385-6390. doi: 10.1021/acs.nanolett.9b02565. Epub 2019 Aug 28. Nano Lett. 2019. PMID: 31438681 Free PMC article.
-
A plasmonic nanorod that walks on DNA origami.Nat Commun. 2015 Aug 25;6:8102. doi: 10.1038/ncomms9102. Nat Commun. 2015. PMID: 26303016 Free PMC article.
-
DNA-Nanotechnology-Enabled Chiral Plasmonics: From Static to Dynamic.Acc Chem Res. 2017 Dec 19;50(12):2906-2914. doi: 10.1021/acs.accounts.7b00389. Epub 2017 Sep 27. Acc Chem Res. 2017. PMID: 28953361
-
DNA nanotechnology for nanophotonic applications.Nanoscale. 2015 Feb 14;7(6):2210-20. doi: 10.1039/c4nr06283c. Nanoscale. 2015. PMID: 25592639 Review.
-
Functional DNA nanostructures for photonic and biomedical applications.Small. 2013 Jul 8;9(13):2210-22. doi: 10.1002/smll.201300141. Epub 2013 Jun 4. Small. 2013. PMID: 23733711 Review.
Cited by
-
A Chirality-Based Quantum Leap.ACS Nano. 2022 Apr 26;16(4):4989-5035. doi: 10.1021/acsnano.1c01347. Epub 2022 Mar 23. ACS Nano. 2022. PMID: 35318848 Free PMC article. Review.
-
Geometry guided crystallization of anisotropic DNA origami shapes.Chem Sci. 2023 Oct 3;14(41):11507-11514. doi: 10.1039/d3sc02722h. eCollection 2023 Oct 25. Chem Sci. 2023. PMID: 37886088 Free PMC article.
-
Self-assembled inorganic chiral superstructures.Nat Rev Chem. 2022 Feb;6(2):125-145. doi: 10.1038/s41570-021-00350-w. Epub 2022 Jan 17. Nat Rev Chem. 2022. PMID: 37117298 Review.
-
Dimerization and oligomerization of DNA-assembled building blocks for controlled multi-motion in high-order architectures.Nat Commun. 2021 May 28;12(1):3207. doi: 10.1038/s41467-021-23532-y. Nat Commun. 2021. PMID: 34050157 Free PMC article.
-
Tunable Chiral Optics in All-Solid-Phase Reconfigurable Dielectric Nanostructures.Nano Lett. 2021 Jan 27;21(2):973-979. doi: 10.1021/acs.nanolett.0c03957. Epub 2020 Dec 29. Nano Lett. 2021. PMID: 33372805 Free PMC article.
References
Publication types
LinkOut - more resources
Full Text Sources

