The Gene Ontology Resource: 20 years and still GOing strong
- PMID: 30395331
- PMCID: PMC6323945
- DOI: 10.1093/nar/gky1055
The Gene Ontology Resource: 20 years and still GOing strong
Abstract
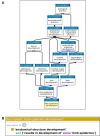
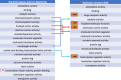
The Gene Ontology resource (GO; http://geneontology.org) provides structured, computable knowledge regarding the functions of genes and gene products. Founded in 1998, GO has become widely adopted in the life sciences, and its contents are under continual improvement, both in quantity and in quality. Here, we report the major developments of the GO resource during the past two years. Each monthly release of the GO resource is now packaged and given a unique identifier (DOI), enabling GO-based analyses on a specific release to be reproduced in the future. The molecular function ontology has been refactored to better represent the overall activities of gene products, with a focus on transcription regulator activities. Quality assurance efforts have been ramped up to address potentially out-of-date or inaccurate annotations. New evidence codes for high-throughput experiments now enable users to filter out annotations obtained from these sources. GO-CAM, a new framework for representing gene function that is more expressive than standard GO annotations, has been released, and users can now explore the growing repository of these models. We also provide the 'GO ribbon' widget for visualizing GO annotations to a gene; the widget can be easily embedded in any web page.
Figures


Similar articles
-
Expansion of the Gene Ontology knowledgebase and resources.Nucleic Acids Res. 2017 Jan 4;45(D1):D331-D338. doi: 10.1093/nar/gkw1108. Epub 2016 Nov 29. Nucleic Acids Res. 2017. PMID: 27899567 Free PMC article.
-
Gene Ontology Consortium: going forward.Nucleic Acids Res. 2015 Jan;43(Database issue):D1049-56. doi: 10.1093/nar/gku1179. Epub 2014 Nov 26. Nucleic Acids Res. 2015. PMID: 25428369 Free PMC article.
-
Large-scale inference of gene function through phylogenetic annotation of Gene Ontology terms: case study of the apoptosis and autophagy cellular processes.Database (Oxford). 2016 Dec 26;2016:baw155. doi: 10.1093/database/baw155. Print 2016. Database (Oxford). 2016. PMID: 28025345 Free PMC article.
-
The Renal Gene Ontology Annotation Initiative.Organogenesis. 2010 Apr-Jun;6(2):71-5. doi: 10.4161/org.6.2.11294. Organogenesis. 2010. PMID: 20885853 Free PMC article. Review.
-
Exploring autophagy with Gene Ontology.Autophagy. 2018;14(3):419-436. doi: 10.1080/15548627.2017.1415189. Epub 2018 Feb 17. Autophagy. 2018. PMID: 29455577 Free PMC article. Review.
Cited by
-
Molecular mechanisms of heterosis under drought stress in maize hybrids Zhengdan7137 and Zhengdan7153.Front Plant Sci. 2024 Oct 8;15:1487639. doi: 10.3389/fpls.2024.1487639. eCollection 2024. Front Plant Sci. 2024. PMID: 39439513 Free PMC article.
-
Axonopathy precedes cell death in ocular damage mediated by blast exposure.Sci Rep. 2021 Jun 3;11(1):11774. doi: 10.1038/s41598-021-90412-2. Sci Rep. 2021. PMID: 34083587 Free PMC article.
-
Subcellular redox responses reveal different Cu-dependent antioxidant defenses between mitochondria and cytosol.Metallomics. 2022 Nov 24;14(11):mfac087. doi: 10.1093/mtomcs/mfac087. Metallomics. 2022. PMID: 36367501 Free PMC article.
-
Drugmonizome and Drugmonizome-ML: integration and abstraction of small molecule attributes for drug enrichment analysis and machine learning.Database (Oxford). 2021 Mar 31;2021:baab017. doi: 10.1093/database/baab017. Database (Oxford). 2021. PMID: 33787872 Free PMC article.
-
Draft Genome Sequence of the Bacterium Paraburkholderia aromaticivorans AR20-38, a Gram-Negative, Cold-Adapted Degrader of Aromatic Compounds.Microbiol Resour Announc. 2020 Jul 2;9(27):e00463-20. doi: 10.1128/MRA.00463-20. Microbiol Resour Announc. 2020. PMID: 32616634 Free PMC article.
References
-
- Fishel R., Ewel A., Lescoe M.K.. Purified human MSH2 protein binds to DNA containing mismatched nucleotides. Cancer Res. 1994; 54:5539–5542. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

