Similar neural responses predict friendship
- PMID: 29382820
- PMCID: PMC5790806
- DOI: 10.1038/s41467-017-02722-7
Similar neural responses predict friendship
Abstract
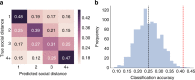
Human social networks are overwhelmingly homophilous: individuals tend to befriend others who are similar to them in terms of a range of physical attributes (e.g., age, gender). Do similarities among friends reflect deeper similarities in how we perceive, interpret, and respond to the world? To test whether friendship, and more generally, social network proximity, is associated with increased similarity of real-time mental responding, we used functional magnetic resonance imaging to scan subjects' brains during free viewing of naturalistic movies. Here we show evidence for neural homophily: neural responses when viewing audiovisual movies are exceptionally similar among friends, and that similarity decreases with increasing distance in a real-world social network. These results suggest that we are exceptionally similar to our friends in how we perceive and respond to the world around us, which has implications for interpersonal influence and attraction.
Conflict of interest statement
The authors declare no competing financial interests.
Figures

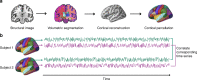
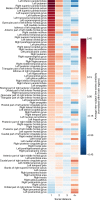
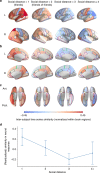

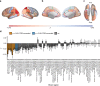
Comment in
-
Birds of a Feather Synchronize Together.Trends Cogn Sci. 2018 May;22(5):371-372. doi: 10.1016/j.tics.2018.03.001. Epub 2018 Mar 13. Trends Cogn Sci. 2018. PMID: 29548666 No abstract available.
Similar articles
-
Social network proximity predicts similar trajectories of psychological states: Evidence from multi-voxel spatiotemporal dynamics.Neuroimage. 2020 Aug 1;216:116492. doi: 10.1016/j.neuroimage.2019.116492. Epub 2019 Dec 28. Neuroimage. 2020. PMID: 31887424
-
Similarity in functional brain connectivity at rest predicts interpersonal closeness in the social network of an entire village.Proc Natl Acad Sci U S A. 2020 Dec 29;117(52):33149-33160. doi: 10.1073/pnas.2013606117. Epub 2020 Dec 14. Proc Natl Acad Sci U S A. 2020. PMID: 33318188 Free PMC article.
-
No evidence for a relationship between social closeness and similarity in resting-state functional brain connectivity in schoolchildren.Sci Rep. 2020 Jul 1;10(1):10710. doi: 10.1038/s41598-020-67718-8. Sci Rep. 2020. PMID: 32612156 Free PMC article.
-
[Recovery of intersubjectivity and empathy in schizophrenics: through a characteristic type of friendship "frolicking"].Seishin Shinkeigaku Zasshi. 2003;105(9):1186-205. Seishin Shinkeigaku Zasshi. 2003. PMID: 14639943 Review. Japanese.
-
Associations between aspects of friendship networks and dietary behavior in youth: Findings from a systematized review.Eat Behav. 2015 Aug;18:7-15. doi: 10.1016/j.eatbeh.2015.03.002. Epub 2015 Mar 20. Eat Behav. 2015. PMID: 25841218 Review.
Cited by
-
Parent-child couples display shared neural fingerprints while listening to stories.Sci Rep. 2024 Feb 4;14(1):2883. doi: 10.1038/s41598-024-53518-x. Sci Rep. 2024. PMID: 38311616 Free PMC article.
-
Shared understanding and social connection: Integrating approaches from social psychology, social network analysis, and neuroscience.Soc Personal Psychol Compass. 2022 Nov;16(11):e12710. doi: 10.1111/spc3.12710. Epub 2022 Oct 17. Soc Personal Psychol Compass. 2022. PMID: 36582415 Free PMC article. Review.
-
Similarity Among Friends Serves as a Social Prior: The Assumption That "Birds of a Feather Flock Together" Shapes Social Decisions and Relationship Beliefs.Pers Soc Psychol Bull. 2024 Jun;50(6):823-840. doi: 10.1177/01461672221140269. Epub 2023 Feb 2. Pers Soc Psychol Bull. 2024. PMID: 36727604 Free PMC article.
-
Imaging Real-Time Tactile Interaction With Two-Person Dual-Coil fMRI.Front Psychiatry. 2020 Apr 28;11:279. doi: 10.3389/fpsyt.2020.00279. eCollection 2020. Front Psychiatry. 2020. PMID: 32411021 Free PMC article.
-
Physiological synchrony predicts observational threat learning in humans.Proc Biol Sci. 2020 May 27;287(1927):20192779. doi: 10.1098/rspb.2019.2779. Epub 2020 May 20. Proc Biol Sci. 2020. PMID: 32429814 Free PMC article.
References
-
- Titelman, G. Random House Dictionary of Popular Proverbs and Sayings (Random House, New York, 1996).
-
- McPherson M, Smith-Lovin L, Cook JM. Birds of a feather: homophily in social networks. Annu. Rev. Sociol. 2001;27:415–444. doi: 10.1146/annurev.soc.27.1.415. - DOI
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

