Actin and Actin-Binding Proteins
- PMID: 26988969
- PMCID: PMC4968159
- DOI: 10.1101/cshperspect.a018226
Actin and Actin-Binding Proteins
Abstract
Organisms from all domains of life depend on filaments of the protein actin to provide structure and to support internal movements. Many eukaryotic cells use forces produced by actin polymerization for their motility, and myosin motor proteins use ATP hydrolysis to produce force on actin filaments. Actin polymerizes spontaneously, followed by hydrolysis of a bound adenosine triphosphate (ATP). Dissociation of the γ-phosphate prepares the polymer for disassembly. This review provides an overview of the properties of actin and shows how dozens of proteins control both the assembly and disassembly of actin filaments. These players catalyze nucleotide exchange on actin monomers, initiate polymerization, promote phosphate dissociation, cap the ends of polymers, cross-link filaments to each other and other cellular components, and sever filaments.
Copyright © 2016 Cold Spring Harbor Laboratory Press; all rights reserved.
Figures





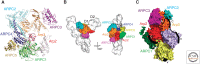
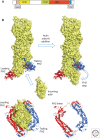
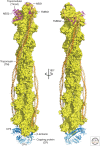

Similar articles
-
Actin polymerization: regulation by divalent metal ion and nucleotide binding, ATP hydrolysis and binding of myosin.Adv Exp Med Biol. 1994;358:71-81. doi: 10.1007/978-1-4615-2578-3_7. Adv Exp Med Biol. 1994. PMID: 7801813 Review.
-
The role of MeH73 in actin polymerization and ATP hydrolysis.J Mol Biol. 2002 Apr 5;317(4):577-89. doi: 10.1006/jmbi.2002.5436. J Mol Biol. 2002. PMID: 11955010
-
Actin polymerization and ATP hydrolysis.Science. 1987 Oct 30;238(4827):638-44. doi: 10.1126/science.3672117. Science. 1987. PMID: 3672117
-
Kinetic evidence for a readily exchangeable nucleotide at the terminal subunit of the barbed ends of actin filaments.Biochemistry. 1998 May 19;37(20):7532-8. doi: 10.1021/bi972176s. Biochemistry. 1998. PMID: 9585568
-
Cellular motility driven by assembly and disassembly of actin filaments.Cell. 2003 Feb 21;112(4):453-65. doi: 10.1016/s0092-8674(03)00120-x. Cell. 2003. PMID: 12600310 Review.
Cited by
-
Reconstituting and Characterizing Actin-Microtubule Composites with Tunable Motor-Driven Dynamics and Mechanics.J Vis Exp. 2022 Aug 25;(186):10.3791/64228. doi: 10.3791/64228. J Vis Exp. 2022. PMID: 36094259 Free PMC article.
-
A computational model to explore how temporal stimulation patterns affect synapse plasticity.PLoS One. 2022 Sep 23;17(9):e0275059. doi: 10.1371/journal.pone.0275059. eCollection 2022. PLoS One. 2022. PMID: 36149886 Free PMC article.
-
Role of Actin-Binding Proteins in Skeletal Myogenesis.Cells. 2023 Oct 25;12(21):2523. doi: 10.3390/cells12212523. Cells. 2023. PMID: 37947600 Free PMC article. Review.
-
Direct observation of the conformational states of formin mDia1 at actin filament barbed ends and along the filament.Mol Biol Cell. 2023 Jan 1;34(1):ar2. doi: 10.1091/mbc.E22-10-0472. Epub 2022 Nov 16. Mol Biol Cell. 2023. PMID: 36383775 Free PMC article.
-
Autoinhibition and relief mechanisms for MICAL monooxygenases in F-actin disassembly.Nat Commun. 2024 Aug 9;15(1):6824. doi: 10.1038/s41467-024-50940-7. Nat Commun. 2024. PMID: 39122694 Free PMC article.
References
-
- Alberts AS. 2001. Identification of a carboxyl-terminal diaphanous-related formin homology protein autoregulatory domain. J Biol Chem 276: 2824–2830. - PubMed
-
- Andrianantoandro E, Pollard TD. 2006. Mechanism of actin filament turnover by severing and nucleation at different concentrations of ADF/cofilin. Mol Cell 24: 13–23. - PubMed
-
- Archer SJ, Vinson VK, Pollard TD, Torchia DA. 1994. Elucidation of the poly-l-proline binding site in Acanthamoeba profilin-I by NMR spectroscopy. FEBS Lett 337: 145–151. - PubMed
-
- Bamburg JR, Harris HE, Weeds AG. 1980. Partial purification and characterization of an actin depolymerizing factor from brain. FEBS Lett 121: 178–182. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous
