Allele-Specific Reprogramming of Cancer Metabolism by the Long Non-coding RNA CCAT2
- PMID: 26853146
- PMCID: PMC4982398
- DOI: 10.1016/j.molcel.2016.01.015
Allele-Specific Reprogramming of Cancer Metabolism by the Long Non-coding RNA CCAT2
Erratum in
- Mol Cell. 2016 Feb 18;61(4):640
Abstract
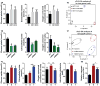
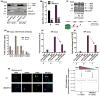
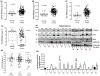
Altered energy metabolism is a cancer hallmark as malignant cells tailor their metabolic pathways to meet their energy requirements. Glucose and glutamine are the major nutrients that fuel cellular metabolism, and the pathways utilizing these nutrients are often altered in cancer. Here, we show that the long ncRNA CCAT2, located at the 8q24 amplicon on cancer risk-associated rs6983267 SNP, regulates cancer metabolism in vitro and in vivo in an allele-specific manner by binding the Cleavage Factor I (CFIm) complex with distinct affinities for the two subunits (CFIm25 and CFIm68). The CCAT2 interaction with the CFIm complex fine-tunes the alternative splicing of Glutaminase (GLS) by selecting the poly(A) site in intron 14 of the precursor mRNA. These findings uncover a complex, allele-specific regulatory mechanism of cancer metabolism orchestrated by the two alleles of a long ncRNA.
Copyright © 2016 Elsevier Inc. All rights reserved.
Figures




Similar articles
-
Molecular Mechanisms for CFIm-Mediated Regulation of mRNA Alternative Polyadenylation.Mol Cell. 2018 Jan 4;69(1):62-74.e4. doi: 10.1016/j.molcel.2017.11.031. Epub 2017 Dec 21. Mol Cell. 2018. PMID: 29276085 Free PMC article.
-
Evidence that cleavage factor Im is a heterotetrameric protein complex controlling alternative polyadenylation.Genes Cells. 2010 Sep 1;15(9):1003-13. doi: 10.1111/j.1365-2443.2010.01436.x. Epub 2010 Jul 29. Genes Cells. 2010. PMID: 20695905
-
Cancer-associated rs6983267 SNP and its accompanying long noncoding RNA CCAT2 induce myeloid malignancies via unique SNP-specific RNA mutations.Genome Res. 2018 Apr;28(4):432-447. doi: 10.1101/gr.225128.117. Epub 2018 Mar 22. Genome Res. 2018. PMID: 29567676 Free PMC article.
-
CCAT2: A novel oncogenic long non-coding RNA in human cancers.Cell Prolif. 2017 Jun;50(3):e12342. doi: 10.1111/cpr.12342. Epub 2017 Feb 28. Cell Prolif. 2017. PMID: 28244168 Free PMC article. Review.
-
Cleavage factor Im (CFIm) as a regulator of alternative polyadenylation.Biochem Soc Trans. 2016 Aug 15;44(4):1051-7. doi: 10.1042/BST20160078. Biochem Soc Trans. 2016. PMID: 27528751 Review.
Cited by
-
Long non-coding RNA colon cancer-associated transcript 2: role and function in human cancers.Chin Med J (Engl). 2022 Dec 5;135(23):2785-2797. doi: 10.1097/CM9.0000000000002286. Chin Med J (Engl). 2022. PMID: 36103972 Free PMC article. Review.
-
Targeting non-coding RNAs to overcome cancer therapy resistance.Signal Transduct Target Ther. 2022 Apr 13;7(1):121. doi: 10.1038/s41392-022-00975-3. Signal Transduct Target Ther. 2022. PMID: 35418578 Free PMC article. Review.
-
Epigenetic regulation of alternative splicing.Am J Cancer Res. 2018 Dec 1;8(12):2346-2358. eCollection 2018. Am J Cancer Res. 2018. PMID: 30662796 Free PMC article. Review.
-
The 'Achilles Heel' of Metabolism in Renal Cell Carcinoma: Glutaminase Inhibition as a Rational Treatment Strategy.Kidney Cancer. 2019 Feb 5;3(1):15-29. doi: 10.3233/KCA-180043. Kidney Cancer. 2019. PMID: 30854496 Free PMC article. Review.
-
Non-Coding Variants in Cancer: Mechanistic Insights and Clinical Potential for Personalized Medicine.Noncoding RNA. 2021 Aug 2;7(3):47. doi: 10.3390/ncrna7030047. Noncoding RNA. 2021. PMID: 34449663 Free PMC article. Review.
References
-
- Cassago A, Ferreira AP, Ferreira IM, Fornezari C, Gomes ER, Greene KS, Pereira HM, Garratt RC, Dias SM, Ambrosio AL. Mitochondrial localization and structure-based phosphate activation mechanism of Glutaminase C with implications for cancer metabolism. Proceedings of the National Academy of Sciences of the United States of America. 2012;109:1092–1097. - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

