A microRNA-30e/mitochondrial uncoupling protein 2 axis mediates TGF-β1-induced tubular epithelial cell extracellular matrix production and kidney fibrosis
- PMID: 23515048
- PMCID: PMC3731557
- DOI: 10.1038/ki.2013.80
A microRNA-30e/mitochondrial uncoupling protein 2 axis mediates TGF-β1-induced tubular epithelial cell extracellular matrix production and kidney fibrosis
Abstract
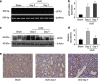
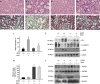
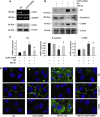
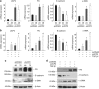
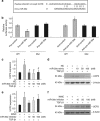
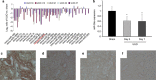
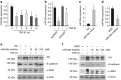
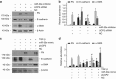
Mitochondria dysfunction has been reported in various kidney diseases but how it leads to kidney fibrosis and how this is regulated is unknown. Here we found that mitochondrial uncoupling protein 2 (UCP2) was induced in kidney tubular epithelial cells after unilateral ureteral obstruction in mice and that mice with ablated UCP2 resisted obstruction-induced kidney fibrosis. We tested this association further in cultured NRK-52E cells and found that TGF-β1 remarkably induced UCP2 expression. Knockdown of UCP2 largely abolished the effect of TGF-β1, whereas overexpression of UCP2 promoted tubular cell phenotype changes. Analysis using a UCP2 mRNA-3'-untranslated region luciferase construct showed that UCP2 mRNA is a direct target of miR-30e. MiR-30e was downregulated in tubular cells from fibrotic kidneys and TGF-β1-treated NRK-52E cells. A miR-30e mimic significantly inhibited TGF-β1-induced tubular-cell epithelial-mesenchymal transition, whereas a miR-30e inhibitor imitated TGF-β1 effects. Finally, genipin, an aglycone UCP2 inhibitor, significantly ameliorated kidney fibrosis in mice. Thus, the miR-30e/UCP2 axis has an important role in mediating TGF-β1-induced epithelial-mesenchymal transition and kidney fibrosis. Targeting this pathway may shed new light for the future of fibrotic kidney disease therapy.
Figures

Similar articles
-
Exosomal miR-21 from tubular cells contributes to renal fibrosis by activating fibroblasts via targeting PTEN in obstructed kidneys.Theranostics. 2021 Aug 2;11(18):8660-8673. doi: 10.7150/thno.62820. eCollection 2021. Theranostics. 2021. PMID: 34522205 Free PMC article.
-
Silencing of the lncRNA TUG1 attenuates the epithelial-mesenchymal transition of renal tubular epithelial cells by sponging miR-141-3p via regulating β-catenin.Am J Physiol Renal Physiol. 2020 Dec 1;319(6):F1125-F1134. doi: 10.1152/ajprenal.00321.2020. Epub 2020 Nov 2. Am J Physiol Renal Physiol. 2020. PMID: 33135476
-
LncRNA GAS5 protects against TGF-β-induced renal fibrosis via the Smad3/miRNA-142-5p axis.Am J Physiol Renal Physiol. 2021 Oct 1;321(4):F517-F526. doi: 10.1152/ajprenal.00085.2021. Epub 2021 Sep 6. Am J Physiol Renal Physiol. 2021. PMID: 34486400
-
The role of TGF-β and epithelial-to mesenchymal transition in diabetic nephropathy.Cytokine Growth Factor Rev. 2011 Jun;22(3):131-9. doi: 10.1016/j.cytogfr.2011.06.002. Epub 2011 Jul 14. Cytokine Growth Factor Rev. 2011. PMID: 21757394 Review.
-
Emerging role of tumor suppressor p53 in acute and chronic kidney diseases.Cell Mol Life Sci. 2022 Aug 9;79(9):474. doi: 10.1007/s00018-022-04505-w. Cell Mol Life Sci. 2022. PMID: 35941392 Free PMC article. Review.
Cited by
-
Insights into non-coding RNAS: biogenesis, function and their potential regulatory roles in acute kidney disease and chronic kidney disease.Mol Cell Biochem. 2024 Aug 7. doi: 10.1007/s11010-024-05083-0. Online ahead of print. Mol Cell Biochem. 2024. PMID: 39110280 Review.
-
Emerging role and the signaling pathways of uncoupling protein 2 in kidney diseases.Ren Fail. 2024 Dec;46(2):2381604. doi: 10.1080/0886022X.2024.2381604. Epub 2024 Aug 1. Ren Fail. 2024. PMID: 39090967 Free PMC article. Review.
-
A Systematic Review and Meta-Analysis of microRNA Profiling Studies in Chronic Kidney Diseases.Noncoding RNA. 2024 May 3;10(3):30. doi: 10.3390/ncrna10030030. Noncoding RNA. 2024. PMID: 38804362 Free PMC article. Review.
-
UCP2 and pancreatic cancer: conscious uncoupling for therapeutic effect.Cancer Metastasis Rev. 2024 Jun;43(2):777-794. doi: 10.1007/s10555-023-10157-4. Epub 2024 Jan 9. Cancer Metastasis Rev. 2024. PMID: 38194152 Free PMC article. Review.
-
MiR-29b modulates DNA methylation in promoter region of miR-130b in mouse model of Diabetic nephropathy.J Diabetes Metab Disord. 2023 May 25;22(2):1105-1115. doi: 10.1007/s40200-023-01208-2. eCollection 2023 Dec. J Diabetes Metab Disord. 2023. PMID: 37975134 Free PMC article.
References
-
- Schnaper HW, Kopp JB. Renal fibrosis. Front Biosci. 2003;8:e68–e86. - PubMed
-
- Rastaldi MP. Epithelial-mesenchymal transition and its implications for the development of renal tubulointerstitial fibrosis. J Nephrol. 2006;19:407–412. - PubMed
-
- Balaban RS, Mandel LJ. Metabolic substrate utilization by rabbit proximal tubule. An NADH fluorescence study. Am J Physiol. 1988;254:F407–F416. - PubMed
-
- Wirthensohn G, Guder WG. Renal substrate metabolism. Physiol Rev. 1986;66:469–497. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Miscellaneous

