Tension induces a base-paired overstretched DNA conformation
- PMID: 22949705
- PMCID: PMC3458322
- DOI: 10.1073/pnas.1213172109
Tension induces a base-paired overstretched DNA conformation
Abstract
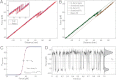
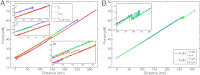
Mixed-sequence DNA molecules undergo mechanical overstretching by approximately 70% at 60-70 pN. Since its initial discovery 15 y ago, a debate has arisen as to whether the molecule adopts a new form [Cluzel P, et al. (1996) Science 271:792-794; Smith SB, Cui Y, Bustamante C (1996) Science 271:795-799], or simply denatures under tension [van Mameren J, et al. (2009) Proc Natl Acad Sci USA 106:18231-18236]. Here, we resolve this controversy by using optical tweezers to extend small 60-64 bp single DNA duplex molecules whose base content can be designed at will. We show that when AT content is high (70%), a force-induced denaturation of the DNA helix ensues at 62 pN that is accompanied by an extension of the molecule of approximately 70%. By contrast, GC-rich sequences (60% GC) are found to undergo a reversible overstretching transition into a distinct form that is characterized by a 51% extension and that remains base-paired. For the first time, results proving the existence of a stretched basepaired form of DNA can be presented. The extension observed in the reversible transition coincides with that produced on DNA by binding of bacterial RecA and human Rad51, pointing to its possible relevance in homologous recombination.
Conflict of interest statement
The authors declare no conflict of interest.
Figures


Similar articles
-
S-DNA and RecA/RAD51-Mediated Strand Exchange in Vitro.Biochemistry. 2019 Apr 16;58(15):2009-2016. doi: 10.1021/acs.biochem.8b01125. Epub 2019 Mar 28. Biochemistry. 2019. PMID: 30900876
-
DNA RECOMBINATION. Base triplet stepping by the Rad51/RecA family of recombinases.Science. 2015 Aug 28;349(6251):977-81. doi: 10.1126/science.aab2666. Science. 2015. PMID: 26315438 Free PMC article.
-
Unraveling the structure of DNA during overstretching by using multicolor, single-molecule fluorescence imaging.Proc Natl Acad Sci U S A. 2009 Oct 27;106(43):18231-6. doi: 10.1073/pnas.0904322106. Epub 2009 Oct 19. Proc Natl Acad Sci U S A. 2009. PMID: 19841258 Free PMC article.
-
Homologous genetic recombination as an intrinsic dynamic property of a DNA structure induced by RecA/Rad51-family proteins: a possible advantage of DNA over RNA as genomic material.Proc Natl Acad Sci U S A. 2001 Jul 17;98(15):8425-32. doi: 10.1073/pnas.111005198. Proc Natl Acad Sci U S A. 2001. PMID: 11459985 Free PMC article. Review.
-
Denaturation transition of stretched DNA.Biochem Soc Trans. 2013 Apr;41(2):639-45. doi: 10.1042/BST20120298. Biochem Soc Trans. 2013. PMID: 23514169 Review.
Cited by
-
The Mechanical Properties of RNA-DNA Hybrid Duplex Stretched by Magnetic Tweezers.Biophys J. 2019 Jan 22;116(2):196-204. doi: 10.1016/j.bpj.2018.12.005. Epub 2018 Dec 13. Biophys J. 2019. PMID: 30635125 Free PMC article.
-
Optical tweezers in single-molecule biophysics.Nat Rev Methods Primers. 2021;1:25. doi: 10.1038/s43586-021-00021-6. Epub 2021 Mar 25. Nat Rev Methods Primers. 2021. PMID: 34849486 Free PMC article.
-
Light-Responsive Polymer Particles as Force Clamps for the Mechanical Unfolding of Target Molecules.Nano Lett. 2018 Apr 11;18(4):2630-2636. doi: 10.1021/acs.nanolett.8b00459. Epub 2018 Mar 28. Nano Lett. 2018. PMID: 29589759 Free PMC article.
-
DNA under Force: Mechanics, Electrostatics, and Hydration.Nanomaterials (Basel). 2015 Feb 25;5(1):246-267. doi: 10.3390/nano5010246. Nanomaterials (Basel). 2015. PMID: 28347009 Free PMC article. Review.
-
Elucidating the Role of Topological Constraint on the Structure of Overstretched DNA Using Fluorescence Polarization Microscopy.J Phys Chem B. 2021 Aug 5;125(30):8351-8361. doi: 10.1021/acs.jpcb.1c02708. Epub 2021 Jul 26. J Phys Chem B. 2021. PMID: 34309392 Free PMC article.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

