Topological domains in mammalian genomes identified by analysis of chromatin interactions
- PMID: 22495300
- PMCID: PMC3356448
- DOI: 10.1038/nature11082
Topological domains in mammalian genomes identified by analysis of chromatin interactions
Abstract
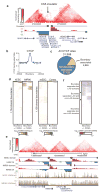
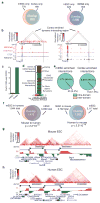
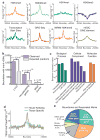
The spatial organization of the genome is intimately linked to its biological function, yet our understanding of higher order genomic structure is coarse, fragmented and incomplete. In the nucleus of eukaryotic cells, interphase chromosomes occupy distinct chromosome territories, and numerous models have been proposed for how chromosomes fold within chromosome territories. These models, however, provide only few mechanistic details about the relationship between higher order chromatin structure and genome function. Recent advances in genomic technologies have led to rapid advances in the study of three-dimensional genome organization. In particular, Hi-C has been introduced as a method for identifying higher order chromatin interactions genome wide. Here we investigate the three-dimensional organization of the human and mouse genomes in embryonic stem cells and terminally differentiated cell types at unprecedented resolution. We identify large, megabase-sized local chromatin interaction domains, which we term 'topological domains', as a pervasive structural feature of the genome organization. These domains correlate with regions of the genome that constrain the spread of heterochromatin. The domains are stable across different cell types and highly conserved across species, indicating that topological domains are an inherent property of mammalian genomes. Finally, we find that the boundaries of topological domains are enriched for the insulator binding protein CTCF, housekeeping genes, transfer RNAs and short interspersed element (SINE) retrotransposons, indicating that these factors may have a role in establishing the topological domain structure of the genome.
Conflict of interest statement
The authors declare no competing financial interests.
Figures

Similar articles
-
Comparative Hi-C reveals that CTCF underlies evolution of chromosomal domain architecture.Cell Rep. 2015 Mar 3;10(8):1297-309. doi: 10.1016/j.celrep.2015.02.004. Epub 2015 Feb 26. Cell Rep. 2015. PMID: 25732821 Free PMC article.
-
Topologically associating domains are stable units of replication-timing regulation.Nature. 2014 Nov 20;515(7527):402-5. doi: 10.1038/nature13986. Nature. 2014. PMID: 25409831 Free PMC article.
-
Insulator function and topological domain border strength scale with architectural protein occupancy.Genome Biol. 2014 Jun 30;15(6):R82. doi: 10.1186/gb-2014-15-5-r82. Genome Biol. 2014. PMID: 24981874 Free PMC article.
-
Chromatin Domains: The Unit of Chromosome Organization.Mol Cell. 2016 Jun 2;62(5):668-80. doi: 10.1016/j.molcel.2016.05.018. Mol Cell. 2016. PMID: 27259200 Free PMC article. Review.
-
Genome-wide studies of CCCTC-binding factor (CTCF) and cohesin provide insight into chromatin structure and regulation.J Biol Chem. 2012 Sep 7;287(37):30906-13. doi: 10.1074/jbc.R111.324962. Epub 2012 Sep 5. J Biol Chem. 2012. PMID: 22952237 Free PMC article. Review.
Cited by
-
Epigenetics and Heart Development.Front Cell Dev Biol. 2021 May 6;9:637996. doi: 10.3389/fcell.2021.637996. eCollection 2021. Front Cell Dev Biol. 2021. PMID: 34026751 Free PMC article. Review.
-
Simultaneous live imaging of the transcription and nuclear position of specific genes.Nucleic Acids Res. 2015 Oct 30;43(19):e127. doi: 10.1093/nar/gkv624. Epub 2015 Jun 19. Nucleic Acids Res. 2015. PMID: 26092696 Free PMC article.
-
Ultrafast and interpretable single-cell 3D genome analysis with Fast-Higashi.Cell Syst. 2022 Oct 19;13(10):798-807.e6. doi: 10.1016/j.cels.2022.09.004. Cell Syst. 2022. PMID: 36265466 Free PMC article.
-
Genomic architecture of autism from comprehensive whole-genome sequence annotation.Cell. 2022 Nov 10;185(23):4409-4427.e18. doi: 10.1016/j.cell.2022.10.009. Cell. 2022. PMID: 36368308 Free PMC article.
-
Domain-Specific and Stage-Intrinsic Changes in Tcrb Conformation during Thymocyte Development.J Immunol. 2015 Aug 1;195(3):1262-72. doi: 10.4049/jimmunol.1500692. Epub 2015 Jun 22. J Immunol. 2015. PMID: 26101321 Free PMC article.
References
-
- Yaffe E, Tanay A. Probabilistic modeling of Hi-C contact maps eliminates systematic biases to characterize global chromosomal architecture. Nat Genet. 43:1059–65. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

