Network motif comparison rationalizes Sec1/Munc18-SNARE regulation mechanism in exocytosis
- PMID: 22423977
- PMCID: PMC3439672
- DOI: 10.1186/1752-0509-6-19
Network motif comparison rationalizes Sec1/Munc18-SNARE regulation mechanism in exocytosis
Abstract
Background: Network motifs, recurring subnetwork patterns, provide significant insight into the biological networks which are believed to govern cellular processes.
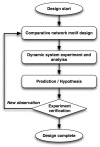
Methods: We present a comparative network motif experimental approach, which helps to explain complex biological phenomena and increases the understanding of biological functions at the molecular level by exploring evolutionary design principles of network motifs.
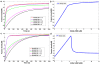
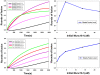
Results: Using this framework to analyze the SM (Sec1/Munc18)-SNARE (N-ethylmaleimide-sensitive factor activating protein receptor) system in exocytic membrane fusion in yeast and neurons, we find that the SM-SNARE network motifs of yeast and neurons show distinct dynamical behaviors. We identify the closed binding mode of neuronal SM (Munc18-1) and SNARE (syntaxin-1) as the key factor leading to mechanistic divergence of membrane fusion systems in yeast and neurons. We also predict that it underlies the conflicting observations in SM overexpression experiments. Furthermore, hypothesis-driven lipid mixing assays validated the prediction.
Conclusion: Therefore this study provides a new method to solve the discrepancies and to generalize the functional role of SM proteins.
Figures



Similar articles
-
The Sec1/Munc18 protein Vps45 regulates cellular levels of its SNARE binding partners Tlg2 and Snc2 in Saccharomyces cerevisiae.PLoS One. 2012;7(11):e49628. doi: 10.1371/journal.pone.0049628. Epub 2012 Nov 14. PLoS One. 2012. PMID: 23166732 Free PMC article.
-
Specific SNARE complex binding mode of the Sec1/Munc-18 protein, Sec1p.Proc Natl Acad Sci U S A. 2006 Nov 21;103(47):17730-5. doi: 10.1073/pnas.0605448103. Epub 2006 Nov 7. Proc Natl Acad Sci U S A. 2006. PMID: 17090679 Free PMC article.
-
The N-peptide-binding mode is critical to Munc18-1 function in synaptic exocytosis.J Biol Chem. 2018 Nov 23;293(47):18309-18317. doi: 10.1074/jbc.RA118.005254. Epub 2018 Oct 1. J Biol Chem. 2018. PMID: 30275014 Free PMC article.
-
At the junction of SNARE and SM protein function.Curr Opin Cell Biol. 2010 Aug;22(4):488-95. doi: 10.1016/j.ceb.2010.04.006. Epub 2010 May 12. Curr Opin Cell Biol. 2010. PMID: 20471239 Free PMC article. Review.
-
Membrane fusion: grappling with SNARE and SM proteins.Science. 2009 Jan 23;323(5913):474-7. doi: 10.1126/science.1161748. Science. 2009. PMID: 19164740 Free PMC article. Review.
Cited by
-
Evolving gene regulatory networks into cellular networks guiding adaptive behavior: an outline how single cells could have evolved into a centralized neurosensory system.Cell Tissue Res. 2015 Jan;359(1):295-313. doi: 10.1007/s00441-014-2043-1. Epub 2014 Nov 23. Cell Tissue Res. 2015. PMID: 25416504 Free PMC article. Review.
-
A computational analysis framework for molecular cell dynamics: case-study of exocytosis.PLoS One. 2012;7(7):e38699. doi: 10.1371/journal.pone.0038699. Epub 2012 Jul 11. PLoS One. 2012. PMID: 22808014 Free PMC article.
-
Munc18-1 controls SNARE protein complex assembly during human sperm acrosomal exocytosis.J Biol Chem. 2012 Dec 21;287(52):43825-39. doi: 10.1074/jbc.M112.409649. Epub 2012 Oct 22. J Biol Chem. 2012. PMID: 23091057 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

