Identification of a cigarette smoke-responsive region in the distal MMP-1 promoter
- PMID: 18617682
- PMCID: PMC2606945
- DOI: 10.1165/rcmb.2007-0310OC
Identification of a cigarette smoke-responsive region in the distal MMP-1 promoter
Abstract
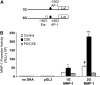
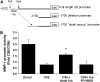
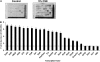
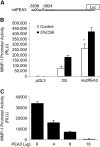
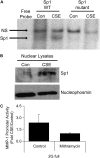
Tobacco-related diseases are leading causes of death worldwide, and many are associated with expression of matrix metalloproteinase-1 (MMP-1). We have reported extracellular signal-regulated kinase (ERK)1/2-dependent induction of MMP-1 by cigarette smoke in lung epithelial cells. Our objectives were to define regions of the human MMP-1 promoter required for activation by smoke, to identify differences in responses of the 1G/2G -1607 polymorphic promoters to smoke, and to identify relevant transcription factors whose activity in airway epithelial cells is increased by smoke. The responses of deletion and mutant promoter constructs were measured in transfected cells during exposure to cigarette smoke extract (CSE). DNA oligonucleotide arrays were used to identify transcription factors activated after smoke exposure. CSE activated the MMP-1 promoter, and this induction was prevented by PD98059 blockade of ERK1/2 phosphorylation. Deletion studies revealed the distal 1kb promoter region (-4438 to -3280 upstream of the transcription start site) is essential for CSE induction of MMP-1, and confers activation of a minimal promoter. Studies of 1G and 2G MMP-1 polymorphic promoter variants revealed higher 2G allele basal and CSE-responsive activities than the 1G allele. Cotransfection, mithramycin, and electrophoretic mobility shift assay studies identified activating and repressive roles for Sp1 and PEA3 transcription factors, respectively. Oligonucleotide DNA arrays confirmed activation of Sp1 and PEA3 by CSE. These data demonstrate that the MMP-1 promoter is a direct target of cigarette smoke in lung epithelial cells. This characterization of a smoke response region in the distal MMP-1 promoter has implications for smoking-related diseases such as cancer, heart disease, and emphysema.
Figures

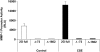


Similar articles
-
The effect of tobacco ingredients on smoke chemistry. Part I: Flavourings and additives.Food Chem Toxicol. 2004;42 Suppl:S3-37. doi: 10.1016/S0278-6915(03)00189-3. Food Chem Toxicol. 2004. PMID: 15072836
-
Tobacco Product Use and Associated Factors Among Middle and High School Students - National Youth Tobacco Survey, United States, 2021.MMWR Surveill Summ. 2022 Mar 11;71(5):1-29. doi: 10.15585/mmwr.ss7105a1. MMWR Surveill Summ. 2022. PMID: 35271557 Free PMC article.
-
Genome-wide methylation profiling and the PI3K-AKT pathway analysis associated with smoking in urothelial cell carcinoma.Cell Cycle. 2013 Apr 1;12(7):1058-70. doi: 10.4161/cc.24050. Epub 2013 Feb 22. Cell Cycle. 2013. PMID: 23435205 Free PMC article.
-
Depressing time: Waiting, melancholia, and the psychoanalytic practice of care.In: Kirtsoglou E, Simpson B, editors. The Time of Anthropology: Studies of Contemporary Chronopolitics. Abingdon: Routledge; 2020. Chapter 5. In: Kirtsoglou E, Simpson B, editors. The Time of Anthropology: Studies of Contemporary Chronopolitics. Abingdon: Routledge; 2020. Chapter 5. PMID: 36137063 Free Books & Documents. Review.
-
Trends in Surgical and Nonsurgical Aesthetic Procedures: A 14-Year Analysis of the International Society of Aesthetic Plastic Surgery-ISAPS.Aesthetic Plast Surg. 2024 Oct;48(20):4217-4227. doi: 10.1007/s00266-024-04260-2. Epub 2024 Aug 5. Aesthetic Plast Surg. 2024. PMID: 39103642 Review.
Cited by
-
The impact of emphysema in pulmonary fibrosis.Eur Respir Rev. 2013 Jun 1;22(128):153-7. doi: 10.1183/09059180.00000813. Eur Respir Rev. 2013. PMID: 23728869 Free PMC article. Review.
-
Validated inference of smoking habits from blood with a finite DNA methylation marker set.Eur J Epidemiol. 2019 Nov;34(11):1055-1074. doi: 10.1007/s10654-019-00555-w. Epub 2019 Sep 7. Eur J Epidemiol. 2019. PMID: 31494793 Free PMC article.
-
Biomarkers Identification in the Microenvironment of Oral Squamous Cell Carcinoma: A Systematic Review of Proteomic Studies.Int J Mol Sci. 2024 Aug 16;25(16):8929. doi: 10.3390/ijms25168929. Int J Mol Sci. 2024. PMID: 39201614 Free PMC article.
-
Site controlled transgenic mice validating increased expression from human matrix metalloproteinase (MMP-1) promoter due to a naturally occurring SNP.Matrix Biol. 2009 Sep;28(7):425-31. doi: 10.1016/j.matbio.2009.06.003. Epub 2009 Jul 3. Matrix Biol. 2009. PMID: 19577645 Free PMC article.
-
Diesel exhaust particles activate the matrix-metalloproteinase-1 gene in human bronchial epithelia in a beta-arrestin-dependent manner via activation of RAS.Environ Health Perspect. 2009 Mar;117(3):400-9. doi: 10.1289/ehp.0800311. Epub 2008 Oct 29. Environ Health Perspect. 2009. PMID: 19337515 Free PMC article.
References
-
- Pardo A, Selman M. MMP-1: the elder of the family. Int J Biochem Cell Biol 2005;37:283–288. - PubMed
-
- Imai K, Dalal S, Chen E, Downey R, Schulman L, Ginsburg M, D'Armiento J. Human collagenase (Matrix Metalloproteinase-1) expression in the lungs of patients with emphysema. Am J Respir Crit Care Med 2001;163:786–791. - PubMed
-
- Biondi ML, Turri O, Leviti S, Seminati R, Cecchini F, Bernini M, Ghilardi G, Guagnellini E. MMP1 and MMP3 polymorphisms in promoter regions and cancer. Clin Chem 2000;46:2023–2024. - PubMed
-
- Osann, KE. Lung cancer in women: the importance of smoking, family history of cancer, and medical history of respiratory disease. Cancer Res 1991;51:4893–4897. - PubMed
-
- Bronnum-Hansen H, Juel K. Estimating mortality due to cigarette smoking: two methods, same result. Epidemiology 2000;11:422–426. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

