Portrait of a species: Chlamydomonas reinhardtii
- PMID: 15956662
- PMCID: PMC1449772
- DOI: 10.1534/genetics.105.044503
Portrait of a species: Chlamydomonas reinhardtii
Abstract
Chlamydomonas reinhardtii, the first alga subject to a genome project, has been the object of numerous morphological, physiological, and genetic studies. The organism has two genetically determined mating types (plus and minus) and all stages of the simple life cycle can be evoked in culture. In the nearly 60 years since the first standard laboratory strains were isolated, numerous crosses and exchanges among laboratories have led to some confusion concerning strain genealogy. Here we use analyses of the nuclear internal transcribed spacer regions and other genetic traits to resolve these issues, correctly identify strains currently available, and analyze phylogenetic relationships with all other available similar chlamydomonad types. The presence of a 10-bp indel in ITS2 in some but not all copies of the nuclear ribosomal cistrons of an individual organism, and the changing ratios of these in crosses, provide a tool to investigate mechanisms of concerted evolution. The standard C. reinhardtii strains, plus C. smithii +, plus the new eastern North American C. reinhardtii isolates, comprise one morphological species, one biological species of high sexual intercompatibility, and essentially identical ITS sequences (except the tip of helix I of ITS2). However, variant RFLP patterns characterize strains from each geographic site.
Figures


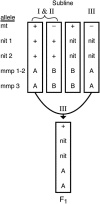

Similar articles
-
Recharacterization of Chlamydomonas reinhardtii and its relatives with new isolates from Japan.J Plant Res. 2010 Jan;123(1):67-78. doi: 10.1007/s10265-009-0266-0. Epub 2009 Oct 31. J Plant Res. 2010. PMID: 19882207
-
Genealogical relationships among laboratory strains of Chlamydomonas reinhardtii as inferred from matrix metalloprotease genes.Curr Genet. 2002 May;41(2):115-22. doi: 10.1007/s00294-002-0284-0. Epub 2002 Apr 19. Curr Genet. 2002. PMID: 12073093
-
Gametogenesis in the Chlamydomonas reinhardtii minus mating type is controlled by two genes, MID and MTD1.Genetics. 2007 Jun;176(2):913-25. doi: 10.1534/genetics.106.066167. Epub 2007 Apr 15. Genetics. 2007. PMID: 17435233 Free PMC article.
-
Chlamydomonas reinhardtii as a new model system for studying the molecular basis of the circadian clock.FEBS Lett. 2011 May 20;585(10):1495-502. doi: 10.1016/j.febslet.2011.02.025. Epub 2011 Feb 25. FEBS Lett. 2011. PMID: 21354416 Review.
-
Evolutionary genetics. The nature and origin of mating types.Curr Biol. 1994 Aug 1;4(8):739-41. doi: 10.1016/s0960-9822(00)00165-2. Curr Biol. 1994. PMID: 7953566 Review.
Cited by
-
Chlamydomonas reinhardtii: A Factory of Nutraceutical and Food Supplements for Human Health.Molecules. 2023 Jan 25;28(3):1185. doi: 10.3390/molecules28031185. Molecules. 2023. PMID: 36770853 Free PMC article. Review.
-
The Chlamydomonas Genome Project, version 6: Reference assemblies for mating-type plus and minus strains reveal extensive structural mutation in the laboratory.Plant Cell. 2023 Feb 20;35(2):644-672. doi: 10.1093/plcell/koac347. Plant Cell. 2023. PMID: 36562730 Free PMC article.
-
A gap-free genome assembly of Chlamydomonas reinhardtii and detection of translocations induced by CRISPR-mediated mutagenesis.Plant Commun. 2023 Mar 13;4(2):100493. doi: 10.1016/j.xplc.2022.100493. Epub 2022 Nov 17. Plant Commun. 2023. PMID: 36397679 Free PMC article.
-
Unassembled cell wall proteins form aggregates in the extracellular space of Chlamydomonas reinhardtii strain UVM4.Appl Microbiol Biotechnol. 2022 Jun;106(11):4145-4156. doi: 10.1007/s00253-022-11960-9. Epub 2022 May 23. Appl Microbiol Biotechnol. 2022. PMID: 35599258 Free PMC article.
-
Systems-wide analysis revealed shared and unique responses to moderate and acute high temperatures in the green alga Chlamydomonas reinhardtii.Commun Biol. 2022 May 13;5(1):460. doi: 10.1038/s42003-022-03359-z. Commun Biol. 2022. PMID: 35562408 Free PMC article.
References
-
- Coleman, A. W., 2000. The significance of a coincidence between evolutionary landmarks found in mating affinity and a DNA sequence. Protist 151: 1–9. - PubMed
-
- Coleman, A. W., 2003. ITS2 is a double-edged tool for eukaryote evolutionary comparisons. Trends Genet. 19: 370–375. - PubMed
-
- Coleman, A. W., and L. J. Goff, 1991. DNA analysis of eukaryote algal species. J. Phycol. 27: 463–473.
-
- Coleman, A. W., and J. C. Mai, 1997. Ribosomal DNA ITS-1 and ITS-2 sequence comparisons as a tool for predicting genetic relatedness. J. Mol. Evol. 45: 168–177. - PubMed
-
- Davies, D. R., and A. Plaskitt, 1971. Genetic and structural analyses of cell-wall formation in Chlamydomonas reinhardi. Genet. Res. 17: 33–43.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

